Research
Methodology
Trained as a statistician, I have long standing interest in statistical methodology development for graphical models, either directed graphs, e.g. directed acyclic graphs (DAG) or undirected graphs, e.g. Gaussian graphical models as they serve a compact and elegant way to decode and visualize complex relations among random variables and thus generate wide applications in many fields, such as social network, biological pathway analysis and Gaussian Copula graphical model in financial market. Most applications center around graphical models. I am also generally interested in developing novel statistical methods(frequentist approach or Bayesian method) driven by problems arising in biostatistics, public health, (epi)genetics and genomics.
Collaboration
As a Biostatistician in our dental school, I am excited to work with faculties, residents and students to support their projects, thesis, etc by providing statistical analysis and consulting. I am also collaborating with researchers on Marquette campus and beyond in data science, public health and statistical genetics.
Graphical models and network analysis
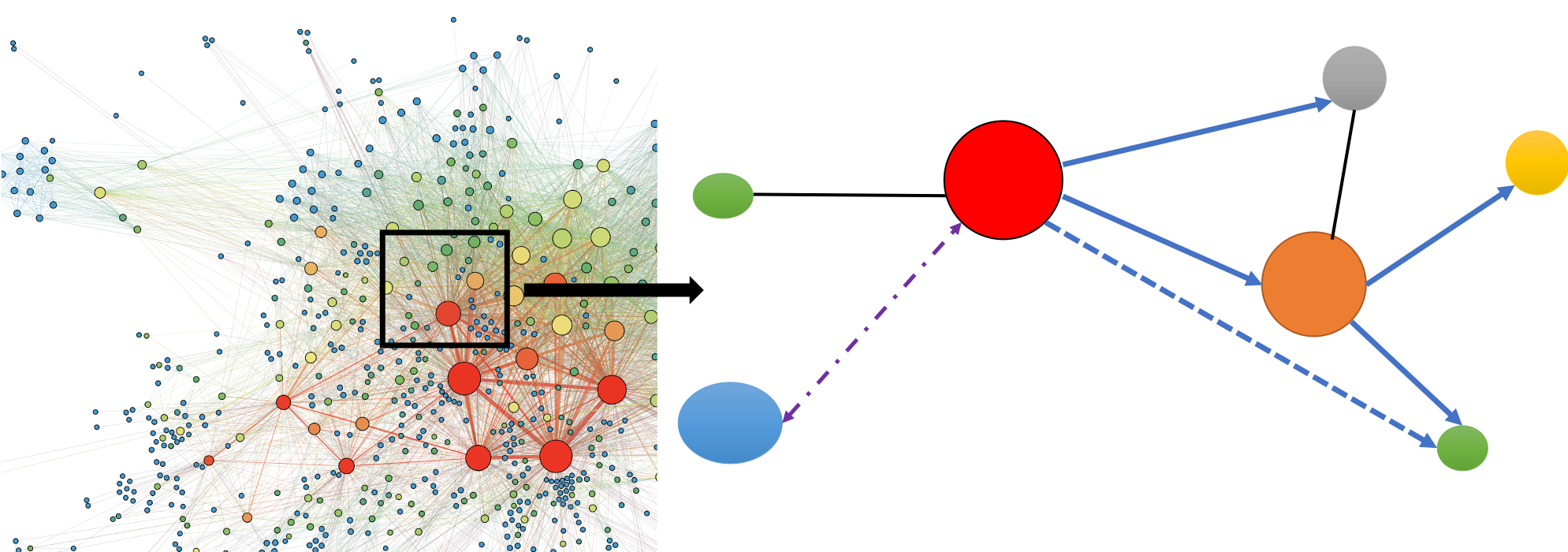
Graphical models have attracted increasing attentions in computer science, machine learning and artificial intelligence. One challenging problem is developing computationally efficient algorithms with nice statistical property to infer graph structure which allows one to identity conditional (in)dependence relations among variables. The computing burden is even more severe for directed graphs as directionality brings complexity in. We are developing novel statistically robust and computationally efficient machine learning algorithms to address these challenges.
1. Gaussian Bayesian network comparisons with graph ordering unknown
Hongmei Zhang, Xianzheng Huang, Shengtong Han, Faisal I. Rezwan, Wilfried Karmaus, Hasan Arshad, John W. Holloway. Computational Statistics and Data Analysis, 2021, 157, 107156. [PDF] [Link]
2. An efficient Bayesian approach for Gaussian Bayesian network structure learning
Shengtong Han, Hongmei Zhang, Wilfried Karmaus, Graham Roberts, Hasan Arshad. Communications in Statistics-Simulation and Computation, 2017, 46, 5070-5084. [PDF][Link]
3. A Full Bayesian Approach for Boolean Genetic Network Inference
Shengtong Han, Raymond K. W. Wong, Thomas C. M. Lee, Linghao Shen, Shuo-Yen R. Li, Xiaodan Fan. PLoS ONE, 2014, 9(12): e115806. doi:10.1371/journal.pone. 0115806. [PDF][Link]
Statistical (epi)genetics and genomics
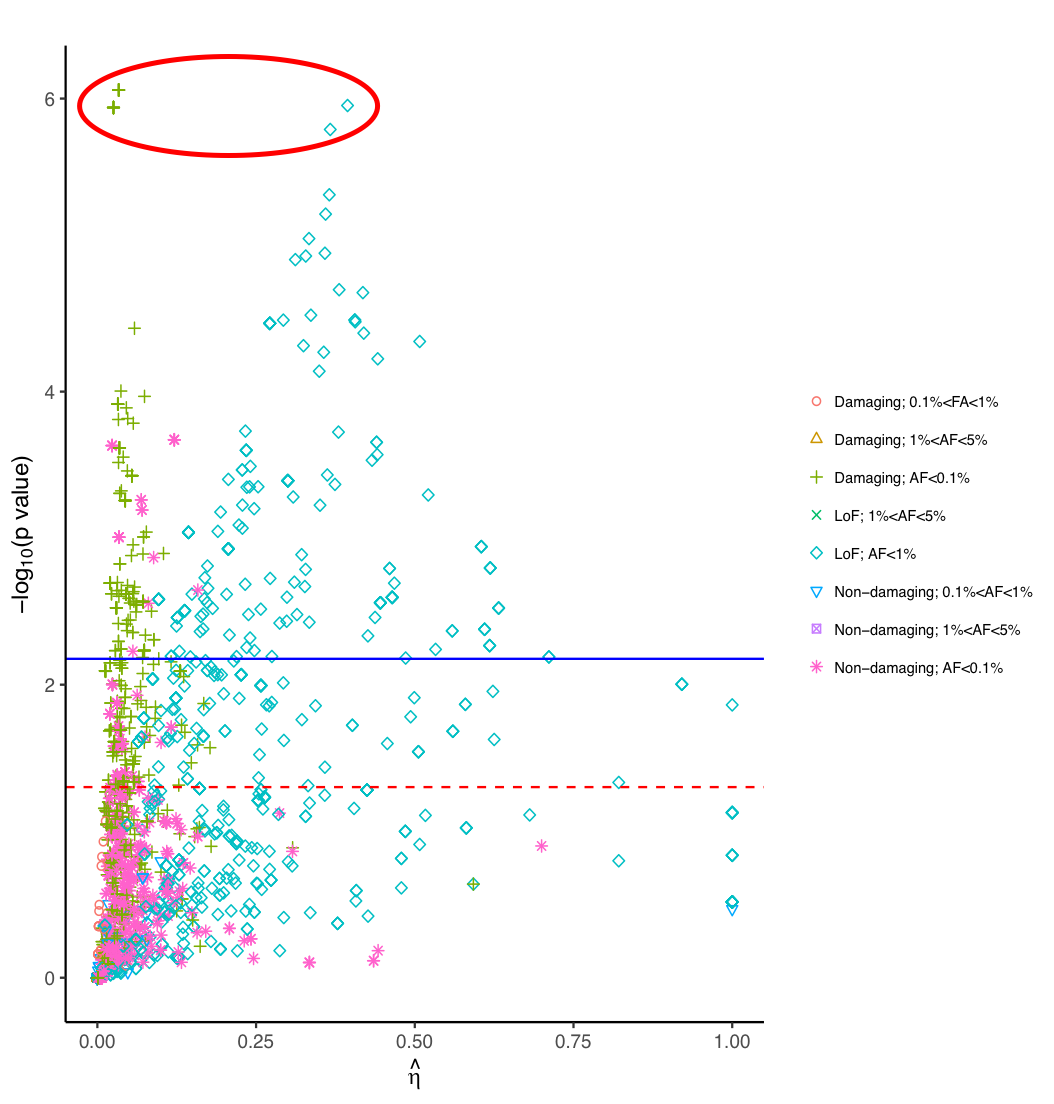
Rare variants (RV) are believed to have large effect size and have the potential to explain missing heritability in human complex traits such as autism. Rare variant association tests tend to be underpowered due to low allele frequency and simple pooling of variants with heterogeneous effect size, e.g. LoF variants tend to be more deleterious than missense variants. We are developing a Bayesian framework to boost the statistical power by accounting for heterogeneous effect size in different variant groups defined based on numerous functional annotations, such as Polyphen, CADD score and apply the method to study the genetics of human complex diseases, such as Autism Spectrum Disorders (ASD) and Alzheimer’s diseases (AD).
1. A Bayesian method for rare variant analysis using functional annotations and its application to Autism
Shengtong Han, Nicholas Knoblauch, Gao Wang, Siming Zhao, Yuwen Liu, Yubin Xie, Wenhui Sheng, Hoang T Nguyen, Xin He. [bioRxiv][R-package]
2. Identifying heterogeneous transgenerational DNA Methylation sites via clustering in Beta regression
Shengtong Han, Hongmei Zhang, Gabrielle A. Lockett, Nandini Mukherjee, John W. Holloway, Wilfried J.J. Karmaus. The Annals of Applied Statistics, 2015, 9, 2052-2072. [PDF][Link]
Computational Biology and Bioinformatics
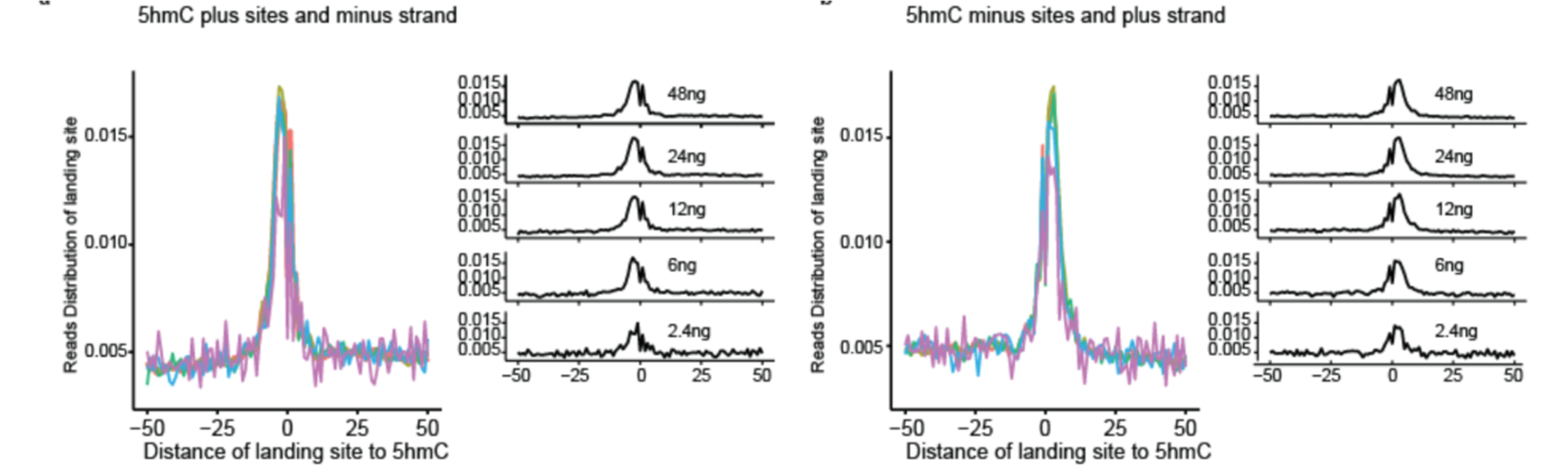
Jump-seq: genome-wide capture and amplification of 5hmC sites
Lulu Hu+, Yuwen Liu+, Shengtong Han+, Lei Yang+, Xiaolong Cui, Yawei Gao, Qing Dai, Xingyu Lu, Xiaochen Kou, Yanhong Zhao, Wenhui Sheng, Shaorong Gao, Xin He and Chuan He. Journal of American Chemical Society, 2019, 141, 22, 8694-8697. [PDF][Link]
